Introduction
Acute Respiratory Distress Syndrome (ARDS) is a common and devastating critical illness. ARDS may be caused by respiratory infections, such as H5N1 avian influenza, or have non-pulmonary pathogenesis, such as septic bacteria or major trauma. Moreover, the COVID-19 pandemic has seen a surge of patients with ARDS in intensive care units across the globe. ARDS is frequently missed or diagnosed late by practicing clinicians (1,2). When patients with ARDS go unrecognized, they do not receive established ARDS treatments and suffer worse outcomes. This article covers data information, experimental setup, and baseline model performance. Future articles will include modeling building using RNN or attention-based neural network.
Objective
The goal is to build a prediction model for ARDS that 1) uses only structured data automatically collected from the EHR and 2) produces estimates of ARDS risk at relevant time points throughout a patient’s course of illness would be of great clinical value. Such models could be applied prospectively with minimal resources to identify ARDS risk and guide therapeutic interventions.
Data Source
The data comes from an anonymous EHR. The data set contains around 9000 distinct id age 18 years or greater with at least one time-stamped assessment each of heart rate, SpO2, blood pressure, creatinine, respiratory rate, temperature, platelet count and WBC count. All subjects in the data were presumably followed routinely and assessed according to the Berlin definition (PaO2/FiO2 < 300). Since the goal was to provide early warnings of ARDS onset, only the structured data elements was used in the analysis as the data could be readily extracted from the EHR in real-time. Thus, the data from unstructured free-text notes or discharge diagnosis codes was not included.
Experimental Setup
Every id in the project was represented by a sequence of events, with each event providing patient information was recorded within a 12 hour period. Only data up to 5 days was used in model building.
Data Preprocessing
Through a series of preprocessing steps, age and gender were mapped to a one-hot or multi-hot vector of binary values (1 or 0). For example, age was first discretized into quantiles of the following ranges: < 20, 20–40, 41–65, 66-80, > 80, then each patient’s age was mapped to a five-dimensional binary feature. Thus, a 66-year old patient would be represented as the five-dimensional feature vector (0, 0, 0, 1, 0).
Other continuous features were transformed into factors(normal: 0, low: -1, high: 1, very low: -2 or very high: 2) to capture potential non-linear relationships, while maintaining the simplicity and interpretability of a linear model.
No imputation of missing numerical values was performed, because explicit imputation of missing values does not always provide consistent improvements to predictive models based on electronic health records (3). Instead, the number -99 was used to associated the missing value to enable models to distinguish between the absence of a numerical value and an actual value of zero.
Baseline Methods
To setup the baseline, the gradient boosted trees (GBTs), logistic regression (LR), random forest (RF), and bayesian logistic regression were employed. The input data of the baseline models includes demographics data, laboratory test results, and vital signs. To avoid leaking future information to the models, I only took the features data before ARDS assessment.
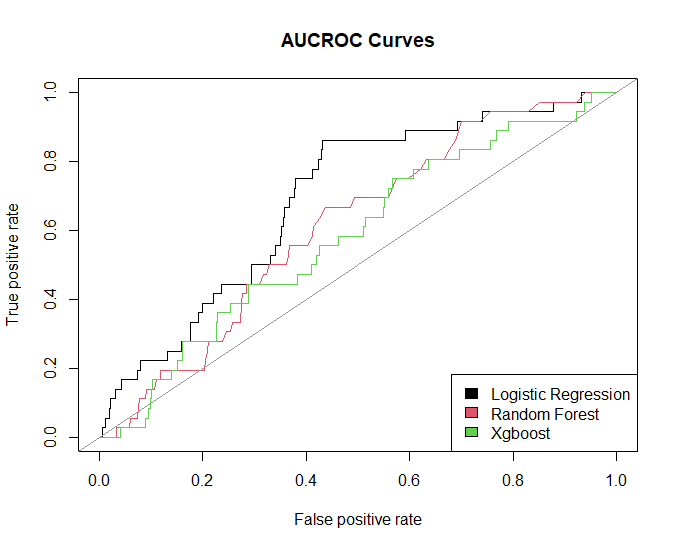
Results
Model evaluation
The test set (total 789, ARDS positive 58) was used to evaluate the performance of learned models (train set , total 2518, ARDS positive 164).
- Confusion matrix:
Leave a comment